Today I want to talk about a paper that looks incredibly cool in a whole bunch of different ways in the abstract, introduction, and discussion but for whom much of that awesomeness falls apart under closer inspection of the results, methods, and context. In it, the authors report their results having flown in NASA’s venerable old DC-8 across the US and down the west coast as well as through a couple of hurricanes with a filter designed to capture bacteria sized particles hanging out the side. They then took the filter and analyzed it with fluorescent dyes and microscopes as well as genomically to see what was there. In their paper they appear to arrive at five different major conclusions: that viable bacterial cells represented on average around 20% of the total particles in the 0.25- to 1-μm diameter range; that 60 to 100% of the 1.5 × 105 cells m−3 they saw were viable; that bacteria are at least two orders of magnitude more abundant than fungal cells in the troposphere; and that fecal coliforms represent a significant amount of the microbiota of hurricanes after landfall. Additionally, what has made the most splash though, is their speculation that because some of the taxa they determined were present by small subunit rRNA sequencing had been shown to metabolize oxalic acid, a major chemical component of clouds, it was plausible that there was active bacterial metabolism happening in the clouds they analyzed. Unfortunately, despite the journal it is published in and glowing praise from excellent blogs like Not Exactly Rocket Science, Climate Central, Wired, Metafilter, and The Scientist, the speculation is pretty foolish and each of these conclusions is either inherently false, actively misleading, or very difficult to support with their data.
 What I’d love to do with the Authors’ platform
What I’d love to do with the Authors’ platform
Here is the paper:
N DeLeon-Rodriguez, TL Lathem, LM Rodriguez-R, et al. Published 2013 in PNAS. doi: 10.1073/pnas.1212089110
The composition and prevalence of microorganisms in the middle-to-upper troposphere (8–15 km altitude) and their role in aerosol-cloud-precipitation interactions represent important, unresolved questions for biological and atmospheric science. In particular, airborne microorganisms above the oceans remain essentially uncharacterized, as most work to date is restricted to samples taken near the Earth’s surface. Here we report on the microbiome of low- and high-altitude air masses sampled onboard the National Aeronautics and Space Administration DC-8 platform during the 2010 Genesis and Rapid Intensification Processes campaign in the Caribbean Sea. The samples were collected in cloudy and cloud-free air masses before, during, and after two major tropical hurricanes, Earl and Karl. Quantitative PCR and microscopy revealed that viable bacterial cells represented on average around 20% of the total particles in the 0.25- to 1-μm diameter range and were at least an order of magnitude more abundant than fungal cells, suggesting that bacteria represent an important and underestimated fraction of micrometer-sized atmospheric aerosols. The samples from the two hurricanes were characterized by significantly different bacterial communities, revealing that hurricanes aerosolize a large amount of new cells. Nonetheless, 17 bacterial taxa, including taxa that are known to use C1–C4 carbon compounds present in the atmosphere, were found in all samples, indicating that these organisms possess traits that allow survival in the troposphere. The findings presented here suggest that the microbiome is a dynamic and underappreciated aspect of the upper troposphere with potentially important impacts on the hydrological cycle, clouds, and climate.
I will start with the most sensational aspect of the paper speculated about by the authors, their assertion that their results indicate it is plausible there is active bacterial life growing in clouds in the troposphere. While the idea that clouds are themselves life forms, ice seeded by cells and cells fed by oxalic acid attracted by the ice, which the authors go to great lengths to speculate on, is very attractive they neglect to mention the temperature readings taken during the flights anywhere in the paper or even supplementary information, which should have pretty much immediately dismissed all of it. The troposphere that the authors were sampling is typically between -50 and -70°C, which approaches the kinds of temperatures I use in my lab to keep bacterial cells in immortal suspended animation. Indeed, even the most extreme psychrophiles don’t grow much below -12°C and even then only really in the presence of a large amount of salt that helps them keep the water they’re in from freezing. The bacteria that they saw may not have been dead exactly when they were sampled, but they certainly weren’t living, which brings us to their next sexy conclusion.
The authors claim that viable bacterial cells represented on average around 20% of the total particles in the 0.25- to 1-μm diameter range and that 60 to 100% of the 1.5 × 105 cells m−3 they saw were viable. This implies that even if the bacteria involved were not actively living, many of them were at least only mostly dead, in a Princess Bride sense. Unlike ordinary humans, bacteria can generally quite happily be frozen in place and exist suspended indefinitely, coming back to life should they thaw in a favorable situation. To determine just how dead the bacteria they were looking at were, the authors used a set of fluorescent dyes sold by Invitrogen that stains DNA green if it is surrounded by an intact membrane, or red if not, on particles they liberated from their filters. However, while the promotional materiel for the various kinds of kits they could have used from their description talk a big game about determining viability, that is not something they can do, at least not for these authors. All that those dyes can measure is whether cells are intact or not, which in some well characterized kinds of systems can be an excellent proxy for whether or not they are viable, but the authors have no idea if it is for them and it is in fact really profoundly unlikely. Even setting aside the likelihood that a significant portion of what they were staining was not DNA to begin with, the upper atmosphere is an incredibly hostile place that one would expect to leave cells a lot more than just mostly dead, even while leaving them intact. Ultraviolet radiation from the sun would kink the DNA of anything up there causing the same kinds of damage as a sunburn, while the dry conditions should dessicate all but the hardiest spores. This is not to say that it isn’t plausible that there truly are viable bacteria up there, but it is to say that the authors do not measure viability in a remotely meaningful way and cannot really contribute to our understanding of how viable exactly those bacteria might be.
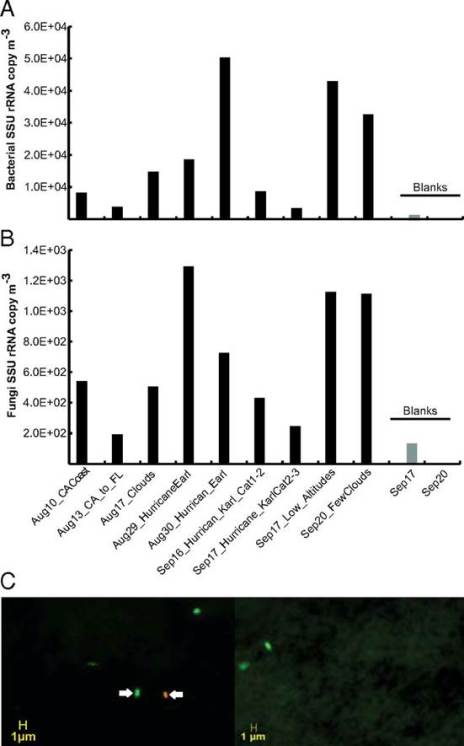
Fig. 1. Quantification of bacterial and fungal cells in samples from high altitudes in the atmosphere. Concentration of bacterial (A) and fungal (B) cells based on qPCR analysis of SSU rRNA gene copies in the samples. Note that samples are ordered by the collection time on the x axis except for blank samples, which are shown at the rightmost part of the graphs in light gray. (C) Live/dead microscopy image of two samples from the California coast and transit flights. Green-stained cells represent cells with viable/intact membrane (e.g., cell indicated by left arrow), and red/yellow-stained cells represent cells with a damaged membrane (e.g., cell indicated by right arrow). Credit: (DeLeon-Rodriguez et al., 2013)
The authors also conclude based on the data shown above that bacteria are at least two orders of magnitude more abundant than fungal cells in the troposphere, and that is indeed what their data plausibly shows. This is to say that even though all of the qPCR data they are relying on is very weak having massive amounts of contamination in their control blanks (just look at the scale), they are blind to that contamination having failed to sequence it, they have no replicates to do statistics with, and they get results that are inconsistent with their other data by two orders of magnitude – they still make a decent case that they see less fungal DNA than bacterial DNA in their filters and that that means fewer fungal cells than bacterial cells. They do however also neglect to mention anywhere in the paper that fungi could be reasonably expected to be less abundant in the late summer when they were measuring, particularly in relation to the early Spring or to a lesser extent mid Fall when fungi tend to sporulate.

Fig. 3. Habitat of origin of the SSU rRNA gene sequences recovered in the GRIP samples. Sequences were assigned to a habitat (see key) based on the source of isolation of their best match in the GreenGenes database. The graph represents the relative abundance of each habitat (vertical axis) for each sample (x axis). Numbers on the top denote the fraction of sequences that were assignable to a habitat for each sample. Credit: (DeLeon-Rodriguez et al., 2013)
The last major finding of this paper, that fecal coliforms represent a significant amount of the microbiota of hurricanes after they make landfall is a really cool one, but if the authors can demonstrate that this is the case, they don’t do it convincingly in this paper. Even setting aside the question of how viable the fecal coliforms they saw were, there is still the very tricky question of the DNA contamination they saw. While I’m sure we can trust that this wasn’t contamination with lab strains from say the autoclave they used for sterilization, which they speculated might be a source in the paper as that would show up very obviously as having little to no diversity, the amount of coliforms they saw is still well within the levels of contamination they know they have. This could all plausibly come from say a livestock operation next to the airport they left from and returned to or some other artifact of the post landfall expeditions they made.

Figure S1. Flight trajectory maps. (A) Flights conducted in the west coast (red) and across the USA (blue). (B) Flights conducted in the area of the Caribbean Sea and the mid-western Atlantic Ocean. The route of each flight is color-coded (see figure key). The trajectory of Hurricane Earl and Karl are colored-coded based on the intensity of the hurricane at each time point (scale bar). Credit: (DeLeon-Rodriguez et al., 2013)
I think it is a particular shame that this research has been sexed up as a microbiota paper that it is not, or at least is not yet, because it still looks like a very interesting climate and meteorological paper. They still are able to convincingly show that that intact bacterial cells in the atmosphere, and particularly within hurricanes, are at least within the same order of magnitude as particles from non-biological origins – this is a really cool and, at least as far as I can tell, novel finding outside of dust from the Sahara. Even if they say silly things about how alive those cells are, have nothing really they could say about how only mostly dead those cells could be, and are limited in what they could say about what kinds of cells they see, that doesn’t mean they couldn’t use stronger techniques in the future. Really to say the kinds of things they want to say, the authors would absolutely need to use culture dependent methods to look at their filters. You just cannot have anything meaningful to say about a bacteria’s ability to grow without actually growing some bacteria. Now, culture dependent methods do have their weaknesses, namely that using them, we only seem to be able to grow about 1% of the bacteria present on Earth, but with the kinds of huge numbers of bacteria the authors are throwing around that shouldn’t scare them. It would also help them to do something I would find really cool: look for viruses.
If they’re actually serious about the contention that there are critters actually actively alive up there, there would be nothing stopping bacteriophages from infecting them, and showing their presence would go a long way to demonstrating that there is ecology going on in clouds. The idea that hurricanes could spread viable cells through the troposphere would go a long way towards explaining why we routinely do things like find in American belly buttons a bacteria that had previously only been found once in Japanese soil; finding viable (by which I strictly mean culturable) bacteriophages in the troposphere would go a long way towards explaining why we see the same thing with them. Now all I need to do is convince the NSF to let me look!
See also a letter to PNAS and its response in the next issue:
Inadequate methods and questionable conclusions in atmospheric life study (Smith & Griffin, 2013)
and
Reply to Smith and Griffin: Methods, air flows, and conclusions are robust in the DeLeon-Rodriguez et al. study (DeLeon-Rodriguez et al., 2013)


 What I’d love to do with the Authors’ platform
What I’d love to do with the Authors’ platform


